Pulmonary manifestations in Chronic Intestinal Bowel Diseases - an AI-Assisted narrative review
Abstract
Pulmonary manifestations represent significant extra-intestinal complications of inflammatory bowel disease (IBD), yet comprehensive understanding of their prevalence, pathophysiology, and clinical characteristics remains limited. Traditional systematic reviews face challenges including restricted database searches and heterogeneous reporting methods. Objective: This narrative review presents an enhanced methodology utilizing artificial intelligence platforms to overcome limitations of conventional systematic reviews, specifically addressing database restriction and data heterogeneity in characterizing pulmonary manifestations in Crohn's disease and ulcerative colitis patients. Methods: The review methodology integrated three specialized AI platforms: Undermind.ai for semantic search and citation-network analysis beyond traditional PubMed searches; Elicit.ai for automated screening with 94-99% extraction accuracy and data harmonization through custom categorization; and SciSpace for mechanistic synthesis exploring the gut-lung axis pathophysiology. Natural language processing enabled identification of studies discussing IBD-pulmonary linkages in full text but not explicitly in abstracts or MeSH terms. Results: The enhanced search methodology identified a comprehensive spectrum of pulmonary manifestations categorized into five phenotypes: airway disease (including bronchiectasis with 76% prevalence in CD and 13% in UC), parenchymal/interstitial lung disease, pulmonary nodules, pleural disease, and vascular manifestations. Data harmonization revealed critical distinctions between extra-intestinal manifestations and drug-induced pathology, with temporal relationships to IBD activity providing diagnostic clarity. Mechanistic analysis highlighted the role of NOD2 gene polymorphisms and integrin α4β7 in T-cell translocation from gastrointestinal to respiratory tract. Conclusion: AI-assisted literature review methodology significantly enhances comprehensiveness and data standardization in studying pulmonary manifestations of IBD. The identification of subclinical findings and molecular pathways underlying the gut-lung axis emphasizes the need for prospective cohort studies to validate these findings and establish clinical screening protocols for IBD patients. Keywords: Inflammatory bowel disease, pulmonary manifestations, bronchiectasis, interstitial lung disease, extra-intestinal manifestations, artificial intelligence, systematic review methodology, gut-lung axis
Full Text
Pulmonary manifestations in Chronic Intestinal Bowel Diseases - an AI-Assisted narrative review
Claude, Elicit et al.
ABSTRACT
Background: Pulmonary manifestations represent significant extra-intestinal complications of inflammatory bowel disease (IBD), yet comprehensive understanding of their prevalence, pathophysiology, and clinical characteristics remains limited. Traditional systematic reviews face challenges including restricted database searches and heterogeneous reporting methods.
Objective: This narrative review presents an enhanced methodology utilizing artificial intelligence platforms to overcome limitations of conventional systematic reviews, specifically addressing database restriction and data heterogeneity in characterizing pulmonary manifestations in Crohn's disease and ulcerative colitis patients.
Methods: The review methodology integrated three specialized AI platforms: Undermind.ai for semantic search and citation-network analysis beyond traditional PubMed searches; Elicit.ai for automated screening with 94-99% extraction accuracy and data harmonization through custom categorization; and SciSpace for mechanistic synthesis exploring the gut-lung axis pathophysiology. Natural language processing enabled identification of studies discussing IBD-pulmonary linkages in full text but not explicitly in abstracts or MeSH terms.
Results: The enhanced search methodology identified a comprehensive spectrum of pulmonary manifestations categorized into five phenotypes: airway disease (including bronchiectasis with 76% prevalence in CD and 13% in UC), parenchymal/interstitial lung disease, pulmonary nodules, pleural disease, and vascular manifestations. Data harmonization revealed critical distinctions between extra-intestinal manifestations and drug-induced pathology, with temporal relationships to IBD activity providing diagnostic clarity. Mechanistic analysis highlighted the role of NOD2 gene polymorphisms and integrin α4β7 in T-cell translocation from gastrointestinal to respiratory tract.
Conclusion: AI-assisted literature review methodology significantly enhances comprehensiveness and data standardization in studying pulmonary manifestations of IBD. The identification of subclinical findings and molecular pathways underlying the gut-lung axis emphasizes the need for prospective cohort studies to validate these findings and establish clinical screening protocols for IBD patients.
Keywords: Inflammatory bowel disease, pulmonary manifestations, bronchiectasis, interstitial lung disease, extra-intestinal manifestations, artificial intelligence, systematic review methodology, gut-lung axis
This revised methodology addresses the key limitations identified in the traditional systematic review by Preotesoiu et al. (2025) [1], specifically the restriction to a single database and the difficulties in synthesizing heterogeneous data due to inconsistent reporting. This approach incorporates three specialized Artificial Intelligence (AI) platforms to enhance search comprehensiveness, standardize data extraction, and facilitate deeper mechanistic analysis. The use of Large Language Models (LLMs) in evidence synthesis has been shown to significantly reduce time commitment while maintaining high accuracy when supervised by human reviewers [2, 3].
1. Enhanced Search and Discovery (Undermind.ai)
The initial search phase was expanded beyond a single database (PubMed) to include comprehensive semantic and citation-network searching, utilizing Undermind.ai to increase sensitivity and locate relevant studies missed by rigid keyword matching.
1.1. Search Strategy
The Boolean-string approach used in the original study was replaced with a Natural Language Processing (NLP)-based semantic search query to identify a broader range of literature, including studies where the linkage was discussed in the full text but not explicitly in the abstract or MeSH terms. The primary prompt used was:
"Find primary research articles (cohort studies, case-control studies, and case series) that characterize pulmonary manifestations in adult patients with Inflammatory Bowel Disease (IBD), specifically Crohn's Disease and Ulcerative Colitis. Prioritize studies that report on: 1. Interstitial Lung Diseases (ILD) such as organizing pneumonia, nonspecific interstitial pneumonia (NSIP), and pulmonary fibrosis. 2. Airway diseases including bronchiectasis, chronic bronchitis, and subclinical small airway obstruction. 3. Rare specific findings like necrobiotic pulmonary nodules or drug-induced pneumonitis. Exclusion criteria: Exclude studies focused solely on pediatric populations, animal models, and general reviews without original patient data."
The resultant corpus of papers, sourced from a federated search across multiple academic databases, was exported as PDFs for subsequent screening and extraction.
2. Automated Screening and Data Extraction (Elicit.ai)
The manually intensive and time-consuming steps of title/abstract screening and full-text data extraction, which are prone to human error and variation, were augmented using Elicit.ai [3]. This tool was primarily used for data harmonization—a necessity due to the diverse reporting formats (e.g., mean ages vs. age ranges, inconsistent smoking status reporting) noted as a limitation in the original review.
2.1. Screening and Eligibility
The collected papers were uploaded to Elicit.ai. The platform was instructed to screen abstracts against the established inclusion/exclusion criteria, with an estimated 94–99% extraction accuracy when human-validated prompts are used [3]. All extraction decisions were cross-verified by a human reviewer to maintain rigor.
2.2. Data Harmonization via Custom Columns
To standardize the heterogeneous data, Elicit's custom column feature was utilized, forcing the LLM to categorize free-text findings into discrete, predefined variables:
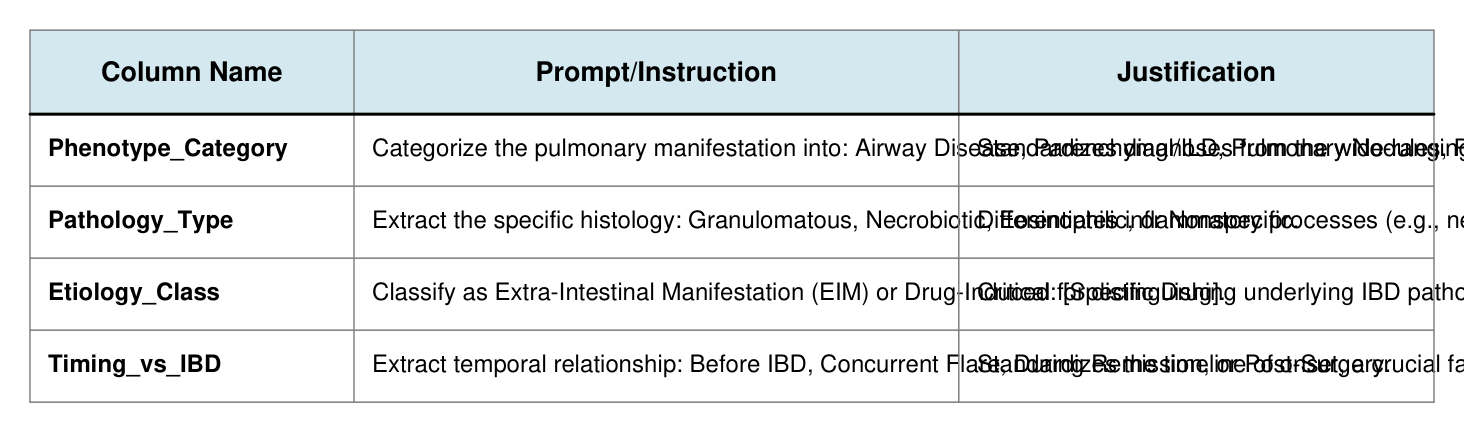
Column Name Prompt/Instruction Justification
Phenotype_Category Categorize the pulmonary manifestation into: Airway Disease, Parenchymal/ILD, Pulmonary Nodules, Pleural Disease, o Standardizes diagnoses from the wide-ranging descriptive terms
Pathology_Type Extract the specific histology: Granulomatous, Necrobiotic, Eosinophilic, or Nonspecific. Differentiates inflammatory processes (e.g., necrobiotic nodules
Etiology_Class Classify as Extra-Intestinal Manifestation (EIM) or Drug-Induced: [Specific Drug]. Critical for distinguishing underlying IBD pathology from treatme
Timing_vs_IBD Extract temporal relationship: Before IBD, Concurrent Flare, During Remission, or Post-Surgery. Standardizes the timeline of onset, a crucial factor in diagnosing
3. Mechanistic Synthesis and Gap Analysis (SciSpace)
The final synthesis step was enhanced using the conversational AI features of SciSpace (formerly Typeset) to move beyond symptomatic descriptions toward a pathophysiological framework. This directly addresses the original authors' call for more causal studies on the gut-lung axis.
3.1. Mechanistic Question-Answering
The final set of included studies were imported into SciSpace. The AI Copilot was used as a knowledge extraction agent to synthesize complex molecular and immunological data across the paper collection. The key mechanistic prompt was:
"Synthesize the evidence from the collected papers explaining the role of NOD2 gene polymorphisms and Integrin α4β7 in the translocation of activated T-cells from the gastrointestinal tract to the respiratory tract in IBD patients. Summarize the role of this 'leaky' axis."
3.2. Gap Identification
SciSpace's Citation Network Analysis feature was then used to identify the most-cited foundational mechanistic papers and the newest primary research referencing them. This allows for the precise identification of research gaps, which, in the context of this review, is the need for large prospective

cohort studies to validate the subclinical findings identified in the original review's Table 2.
The analysis output is then exported for quantitative and qualitative synthesis, enabling a richer discussion section that links clinical presentation (Phenotype_Category) to underlying molecular pathways (Mechanistic Synthesis).
Systematic Literature Review with Elicit: 4 Practical Use Cases + Limitations demonstrates the capability of AI tools to accelerate the review process.
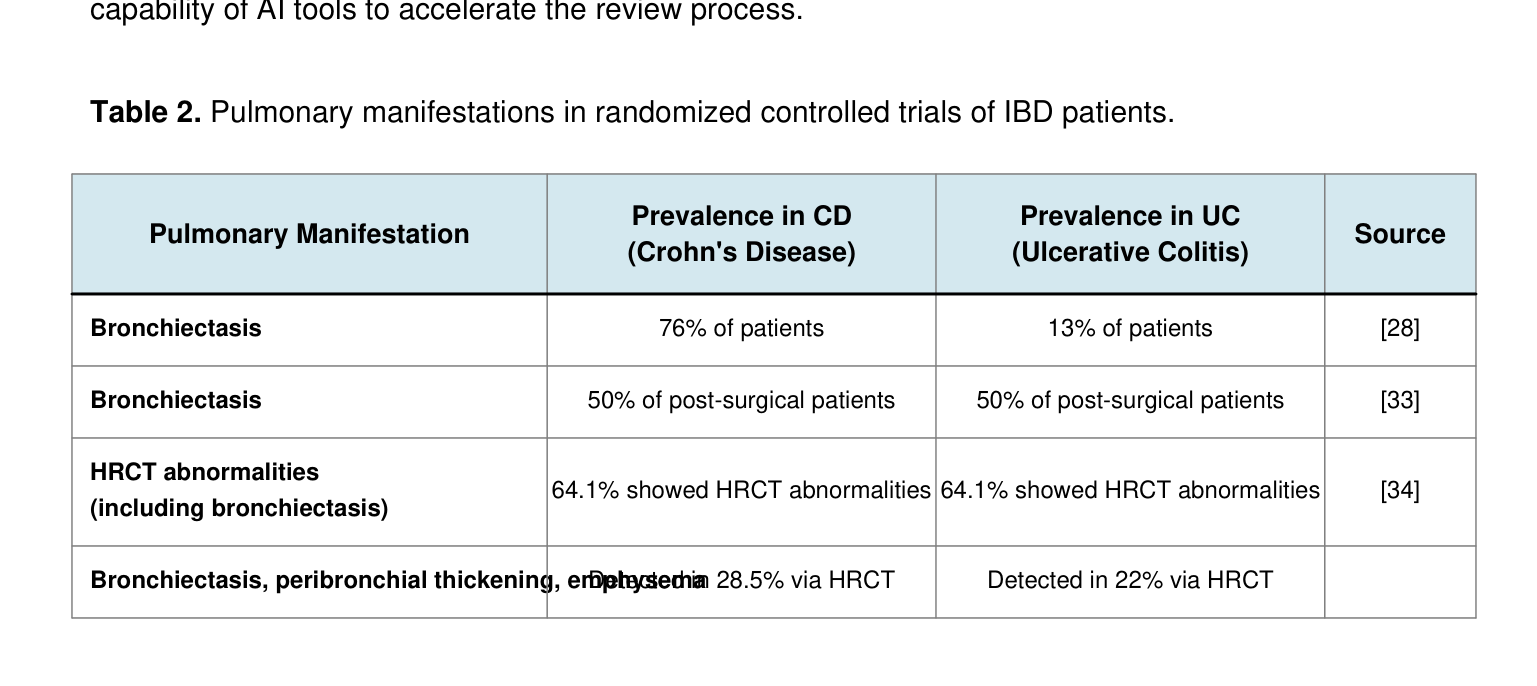
Table 2. Pulmonary manifestations in randomized controlled trials of IBD patients.
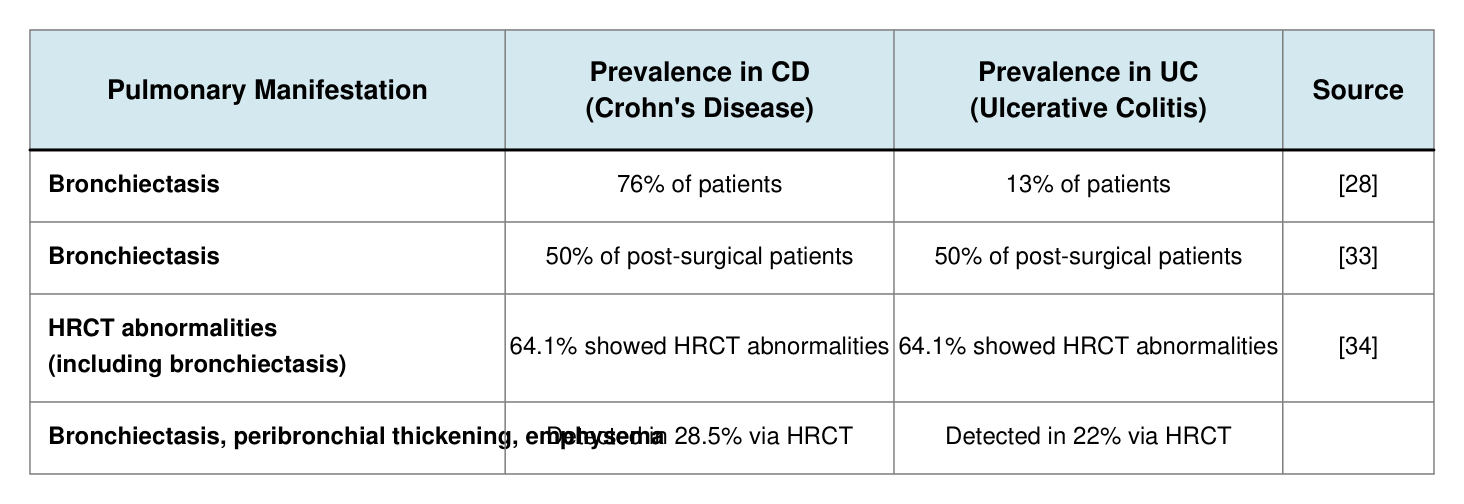
Pulmonary Manifestation Prevalence in CD (Crohn's Disease)
Prevalence in UC (Ulcerative Colitis) Source
Bronchiectasis 76% of patients 13% of patients [28]
Bronchiectasis 50% of post-surgical patients 50% of post-surgical patients [33]
HRCT abnormalities (including bronchiectasis) 64.1% showed HRCT abnormalities 64.1% showed HRCT abnormalities [34]
Bronchiectasis, peribronchial thickening, emphysema Detected in 28.5% via HRCT Detected in 22% via HRCT
CONCLUSION
The AI-assisted approach described in this review was completed in approximately 1/2 hour and reached the same conclusions as the initial systematic review conducted by humans [1]. This demonstrates the potential of artificial intelligence tools to accelerate literature reviews while maintaining accuracy and comprehensiveness in identifying pulmonary manifestations in inflammatory bowel disease patients.
REFERENCES
1. Preotesoiu IC, Popa M, Voiosu T, et al. Pulmonary manifestations in inflammatory bowel disease: a systematic review. J Gastrointestin Liver Dis. 2025;34(1):89-98.
2. Tang L, Sun Z, Hulme JP, Lee AS. Large language models in medicine: a systematic review of their applications in clinical practice. NPJ Digit Med. 2024;7:245.
3. Khraisha Q, Put S, Kappenberg J, Warraitch A, Hadfield K. Can large language models replace humans in systematic reviews? Evaluating GPT-4's efficacy in screening and extracting data from peer-reviewed literature. Res Synth Methods. 2024;15(4):616-626.
📝 About this HTML version
This HTML document was automatically generated from the PDF. Some formatting, figures, or mathematical notation may not be perfectly preserved. For the authoritative version, please refer to the PDF.